Interfacing pytorch models with collections of AnnData objects#
Author: Sergei Rybakov
This tutorial uses
AnnLoaderin combination withAnnCollectionto interface pytorch models, aimed at training models at the scale of many AnnData objects.For basic usage of the AnnLoader and AnnCollection classes, please see their respective tutorials.
import gdown
import torch
import torch.nn as nn
import pyro
import pyro.distributions as dist
import numpy as np
import scanpy as sc
import pandas as pd
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
from anndata.experimental.multi_files import AnnCollection
from anndata.experimental.pytorch import AnnLoader
import warnings
warnings.simplefilter(action="ignore", category=pd.core.common.SettingWithCopyWarning)
VAE model definition#
class MLP(nn.Module):
def __init__(self, input_dim, hidden_dims, out_dim):
super().__init__()
modules = []
for in_size, out_size in zip([input_dim]+hidden_dims, hidden_dims):
modules.append(nn.Linear(in_size, out_size))
modules.append(nn.LayerNorm(out_size))
modules.append(nn.ReLU())
modules.append(nn.Dropout(p=0.05))
modules.append(nn.Linear(hidden_dims[-1], out_dim))
self.fc = nn.Sequential(*modules)
def forward(self, *inputs):
shape = dist.util.broadcast_shape(*[s.shape[:-1] for s in inputs]) + (-1,)
inputs = [s.expand(shape) for s in inputs]
input_cat = torch.cat(inputs, dim=-1)
return self.fc(input_cat)
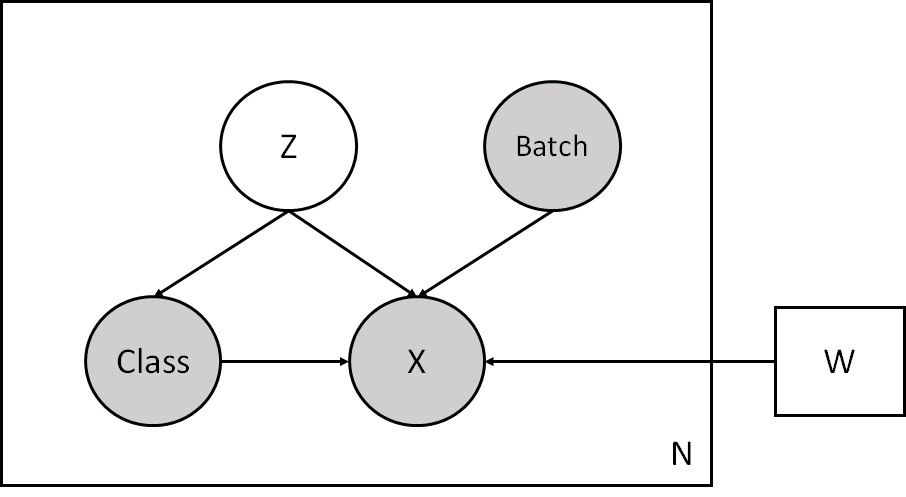
This model is a bit different from the model in the AnnLoader tutorial. Here the counts variable X depends on the class labels Class not only through the latent variable Z but also directly.This makes this model semi-supervised.
# The code is based on the scarches trvae model
# https://github.com/theislab/scarches/blob/v0.3.5/scarches/models/trvae/trvae.py
# and on the pyro.ai Variational Autoencoders tutorial
# http://pyro.ai/examples/vae.html
# and the Semi-Supervised VAE tutorial
# http://pyro.ai/examples/ss-vae.html
class CVAE(nn.Module):
def __init__(self, input_dim, n_conds, n_classes, hidden_dims, latent_dim, classifier_dims=[128]):
super().__init__()
self.encoder = MLP(input_dim+n_conds, hidden_dims, 2*latent_dim) # output - mean and logvar of z
self.decoder = MLP(latent_dim+n_conds+n_classes, hidden_dims[::-1], input_dim)
self.theta = nn.Linear(n_conds, input_dim, bias=False)
self.classifier = MLP(latent_dim, classifier_dims, n_classes)
self.latent_dim = latent_dim
def model(self, x, batches, classes, size_factors, supervised):
pyro.module("cvae", self)
batch_size = x.shape[0]
with pyro.plate("data", batch_size):
z_loc = x.new_zeros((batch_size, self.latent_dim))
z_scale = x.new_ones((batch_size, self.latent_dim))
z = pyro.sample("Z", dist.Normal(z_loc, z_scale).to_event(1))
classes_probs = self.classifier(z).softmax(dim=-1)
if supervised:
obs = classes
else:
obs = None
classes = pyro.sample("Class", dist.OneHotCategorical(probs=classes_probs), obs=obs)
dec_mu = self.decoder(z, batches, classes).softmax(dim=-1) * size_factors[:, None]
dec_theta = torch.exp(self.theta(batches))
logits = (dec_mu + 1e-6).log() - (dec_theta + 1e-6).log()
pyro.sample("X", dist.NegativeBinomial(total_count=dec_theta, logits=logits).to_event(1), obs=x.int())
def guide(self, x, batches, classes, size_factors, supervised):
batch_size = x.shape[0]
with pyro.plate("data", batch_size):
z_loc_scale = self.encoder(x, batches)
z_mu = z_loc_scale[:, :self.latent_dim]
z_var = torch.sqrt(torch.exp(z_loc_scale[:, self.latent_dim:]) + 1e-4)
z = pyro.sample("Z", dist.Normal(z_mu, z_var).to_event(1))
if not supervised:
classes_probs = self.classifier(z).softmax(dim=-1)
pyro.sample("Class", dist.OneHotCategorical(probs=classes_probs))
Create AnnCollection from two AnnData objects#
The data is from this scvi reproducibility notebook.
gdown.download(url="https://drive.google.com/uc?id=1X5N9rOaIqiGxZRyr1fyZ6NpDPeATXoaC",
output="pbmc_seurat_v4.h5ad", quiet=False)
gdown.download(url="https://drive.google.com/uc?id=1JgaXNwNeoEqX7zJL-jJD3cfXDGurMrq9",
output="covid_cite.h5ad", quiet=False)
Downloading...
From: https://drive.google.com/uc?id=1X5N9rOaIqiGxZRyr1fyZ6NpDPeATXoaC
To: C:\Users\sergei.rybakov\projects\anndata-tutorials\pbmc_seurat_v4.h5ad
100%|█████████████████████████████████████████████████████████████████████████████| 1.00G/1.00G [00:47<00:00, 21.3MB/s]
Downloading...
From: https://drive.google.com/uc?id=1JgaXNwNeoEqX7zJL-jJD3cfXDGurMrq9
To: C:\Users\sergei.rybakov\projects\anndata-tutorials\covid_cite.h5ad
100%|███████████████████████████████████████████████████████████████████████████████| 304M/304M [00:13<00:00, 22.2MB/s]
'covid_cite.h5ad'
covid = sc.read('covid_cite.h5ad')
pbmc = sc.read('pbmc_seurat_v4.h5ad')
covid
AnnData object with n_obs × n_vars = 57669 × 33538
obs: 'orig.ident', 'nCount_RNA', 'nFeature_RNA', 'RNA_snn_res.0.4', 'seurat_clusters', 'set', 'Resp', 'disease', 'subj_code', 'covidpt_orhealth', 'mito', 'ncount', 'nfeat', 'bust_21', 'og_clust', 'severmod_other', 'og_clusts', 'nCount_ADT', 'nFeature_ADT', 'UMAP1', 'UMAP2', 'final_clust', 'final_clust_v2', 'new_pt_id', 'Resp_og', 'final_clust_withnum', 'final_clust_review', 'Age', 'Gender', 'Gender_num'
obsm: 'pro_exp'
pbmc
AnnData object with n_obs × n_vars = 161764 × 20729
obs: 'nCount_ADT', 'nFeature_ADT', 'nCount_RNA', 'nFeature_RNA', 'orig.ident', 'lane', 'donor', 'time', 'celltype.l1', 'celltype.l2', 'celltype.l3', 'Phase', 'nCount_SCT', 'nFeature_SCT', 'X_index'
obsm: 'protein_counts'
sc.pp.highly_variable_genes(
pbmc,
n_top_genes=4000,
flavor="seurat_v3",
batch_key="orig.ident",
subset=True,
)
dataset = AnnCollection(
{'covid': covid, 'pbmc':pbmc},
join_vars='inner',
join_obs='outer',
join_obsm='inner',
label='dataset',
indices_strict=False
)
dataset
AnnCollection object with n_obs × n_vars = 219433 × 4000
constructed from 2 AnnData objects
obs: 'orig.ident', 'nCount_RNA', 'nFeature_RNA', 'RNA_snn_res.0.4', 'seurat_clusters', 'set', 'Resp', 'disease', 'subj_code', 'covidpt_orhealth', 'mito', 'ncount', 'nfeat', 'bust_21', 'og_clust', 'severmod_other', 'og_clusts', 'nCount_ADT', 'nFeature_ADT', 'UMAP1', 'UMAP2', 'final_clust', 'final_clust_v2', 'new_pt_id', 'Resp_og', 'final_clust_withnum', 'final_clust_review', 'Age', 'Gender', 'Gender_num', 'lane', 'donor', 'time', 'celltype.l1', 'celltype.l2', 'celltype.l3', 'Phase', 'nCount_SCT', 'nFeature_SCT', 'X_index', 'dataset'
dataset.obs['orig.ident'].loc[covid.obs_names] = covid.obs['set']
dataset.obs['size_factors'] = pd.Series(dtype='float32')
for batch, idx in dataset.iterate_axis(5000):
dataset.obs['size_factors'].iloc[idx] = np.ravel(batch.X.sum(1))
encoder_study = OneHotEncoder(sparse=False, dtype=np.float32)
encoder_study.fit(dataset.obs['orig.ident'].to_numpy()[: , None])
OneHotEncoder(dtype=<class 'numpy.float32'>, sparse=False)
labels = 'celltype.l1'
encoder_celltype = OneHotEncoder(sparse=False, dtype=np.float32)
encoder_celltype.fit(dataset.obs[labels].to_numpy()[: , None])
OneHotEncoder(dtype=<class 'numpy.float32'>, sparse=False)
use_cuda = torch.cuda.is_available()
encoders = {
'obs': {
'orig.ident': lambda s: encoder_study.transform(s.to_numpy()[: , None]),
labels: lambda c: encoder_celltype.transform(c.to_numpy()[: , None])[:, :-1]
}
}
dataset.convert = encoders
AnnLoader and model initialization#
dataloader = AnnLoader(dataset, batch_size=256, shuffle=True, use_cuda=use_cuda)
n_conds = len(encoder_study.categories_[0])
n_classes = len(encoder_celltype.categories_[0])-1
latent_dim = 20
cvae = CVAE(dataset.shape[1], n_conds=n_conds, n_classes=n_classes, hidden_dims=[1280, 256], latent_dim=latent_dim)
if use_cuda:
cvae.cuda()
Train the model#
optimizer = pyro.optim.Adam({"lr": 1e-3})
svi_sup = pyro.infer.SVI(cvae.model, cvae.guide,
optimizer, loss=pyro.infer.TraceMeanField_ELBO())
# very slow for sequential
svi_unsup = pyro.infer.SVI(cvae.model, pyro.infer.config_enumerate(cvae.guide, 'parallel', expand=True),
optimizer, loss=pyro.infer.TraceEnum_ELBO())
def train(svi_sup, svi_unsup, train_loader):
epoch_loss = 0.
num_correct = 0
for batch in train_loader:
# select indices of labelled cells
sup_idx = batch.obs[labels].sum(1).bool().cpu().numpy()
batch_sup = batch[sup_idx]
batch_unsup = batch[~sup_idx]
# do supervised step for the labelled cells
sup_data = batch_sup.obs.to_dict(keys=['orig.ident', labels, 'size_factors'])
sup_data['supervised'] = True
epoch_loss += svi_sup.step(batch_sup.X, *sup_data.values())
# do unsupervised step for unlabelled cells
unsup_data = batch_unsup.obs.to_dict(keys=['orig.ident', labels, 'size_factors'])
unsup_data['supervised'] = False
epoch_loss += svi_unsup.step(batch_unsup.X, *unsup_data.values())
normalizer_train = len(train_loader.dataset)
total_epoch_loss_train = epoch_loss / normalizer_train
return total_epoch_loss_train
NUM_EPOCHS = 250
for epoch in range(NUM_EPOCHS):
total_epoch_loss_train = train(svi_sup, svi_unsup, dataloader)
if epoch % 40 == 0 or epoch == NUM_EPOCHS-1:
print("[epoch %03d] average training loss: %.4f" % (epoch, total_epoch_loss_train))
[epoch 000] average training loss: 983.3127
[epoch 040] average training loss: 898.7591
[epoch 080] average training loss: 895.9434
[epoch 120] average training loss: 894.6037
[epoch 160] average training loss: 893.6797
[epoch 200] average training loss: 893.0895
[epoch 240] average training loss: 892.6837
[epoch 249] average training loss: 892.6027
Check the results#
Get means of latent variables and cell labels predictions for the unlabelled data.
dataset.obsm['X_cvae'] = np.empty((dataset.shape[0], latent_dim), dtype='float32')
dataset.obs['cell_type_pred'] = pd.Series(dtype=pbmc.obs[labels].dtype)
for batch, idx in dataloader.dataset.iterate_axis(5000):
latents = cvae.encoder(batch.X, batch.obs['orig.ident'])[:, :latent_dim].detach()
predict = cvae.classifier(latents).detach()
one_hot = torch.zeros(predict.shape[0], predict.shape[1]+1, device=predict.device)
one_hot = one_hot.scatter_(1, predict.argmax(dim=-1, keepdim=True) , 1.).cpu().numpy()
dataset.obsm['X_cvae'][idx] = latents.cpu().numpy()
dataset.obs['cell_type_pred'].iloc[idx] = np.ravel(enc_ct.inverse_transform(one_hot))
dataset.obs['cell_type_joint'] = dataset.obs['cell_type_pred']
dataset.obs['cell_type_joint'].loc[pbmc.obs_names]=pbmc.obs[labels]
adata = dataset.to_adata()
sc.pp.neighbors(adata, use_rep='X_cvae')
sc.tl.umap(adata)
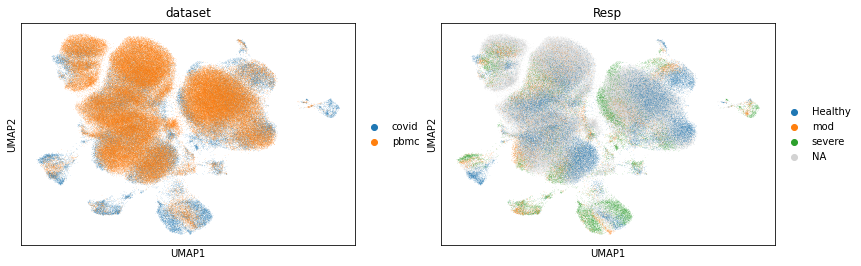
sc.pl.umap(adata, color=['dataset', 'Resp'])
... storing 'orig.ident' as categorical
... storing 'RNA_snn_res.0.4' as categorical
... storing 'seurat_clusters' as categorical
... storing 'set' as categorical
... storing 'Resp' as categorical
... storing 'disease' as categorical
... storing 'subj_code' as categorical
... storing 'covidpt_orhealth' as categorical
... storing 'bust_21' as categorical
... storing 'og_clust' as categorical
... storing 'severmod_other' as categorical
... storing 'og_clusts' as categorical
... storing 'final_clust' as categorical
... storing 'final_clust_v2' as categorical
... storing 'new_pt_id' as categorical
... storing 'Resp_og' as categorical
... storing 'final_clust_withnum' as categorical
... storing 'final_clust_review' as categorical
... storing 'Gender' as categorical
... storing 'lane' as categorical
... storing 'donor' as categorical
... storing 'time' as categorical
... storing 'celltype.l1' as categorical
... storing 'celltype.l2' as categorical
... storing 'celltype.l3' as categorical
... storing 'Phase' as categorical
... storing 'X_index' as categorical
... storing 'dataset' as categorical
accuracy = (dataset.obs['cell_type_pred'].loc[pbmc.obs_names]==pbmc.obs[labels]).sum().item()/pbmc.n_obs
accuracy
0.9234131203481615
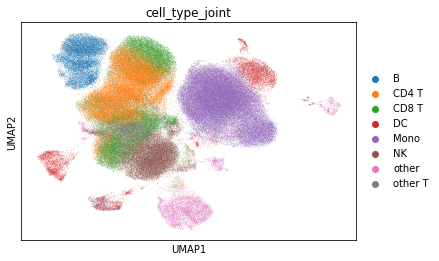
Plot pbmc cell types and predicted cell types for covid.
sc.pl.umap(adata, color='cell_type_joint')